- Author(s): Derin Mary Thomas and A. S. Arun Prasad
- Category: HTML FullText
- Published: 02 January 2017
- Views: 1979
Direct Orange 26 - Azo Dye: Decolorization and Degradation by Bacillus mojavensis ABR
Derin Mary Thomasa and A. S. Arun Prasada,*
aDivision of Environmental Biotechnology, School of Biosciences and Technology, VIT University, Vellore, India.
Published Online 02 01 2017
Abstract:
The potential of the bacterial strain surviving in the harsh conditions of the textile effluents was tested to decolorize and degrade the structurally complex azo dye. The bacterium Bacillus mojavensis strain ABR isolated from the textile effluent was evaluated in decolorizing the azo dye Direct Orange 26 (DO-6). 100 mgl-1 of DO-26 was decolorized with 95% efficiency within 6 hrs at pH 7 and 37°C with 5% NaCl under static conditions in decolorizing medium. Decrease in absorbance at 493 nm (λmax) was periodically evaluated using UV-Vis spectrophotometer to check the decolorization. HPLC showed the formation of metabolites at different retention times indicating degradation of DO-26 which was further confirmed by LC-MS. The less toxic nature of the DO-26 degraded metabolites was confirmed by the phytotoxicity test on seeds of Vigna mungo, Sorghum bicolor and Vigna radiata.
INTRODUCTION
The natural environment we live in is composed of atmosphere, earth, water and space. The composition and complex nature of this stable environment was altered as a result of various human activities such as globalization and industrialization resulting in the setting up of different industries viz. textile industry, paper industry, food industry etc. Among these industries the textile industry is one of the largest and oldest sectors. The textile industry has major impact on nation’s economy as well as on the environmental quality of life.1 The textile industry uses a wide variety of chemical products which contain dispersants, alkalis, organic surfactants, solvents, humectants, acids, salts and residual dyes.2 The textile industry processes consume large amounts of freshwater and release equal quantity of wastewater, making it one of the main sources of severe pollution worldwide.3 The textile wastewaters is rated as the most polluted of all industries and are characterized by fluctuations in many parameters such as pH, temperature, chemical oxygen demand (COD), biological oxygen demand (BOD), salinity and colour.4,5 The wastewaters contain detergents, metals, surfactants, suspended and dissolved solids, dispersants, levelling agents, toxic organics (phenols), chlorinated compounds (AOX) and formaldehyde.6 The most common units of textile processing include desizing, bleaching, mercerising and dyeing processes. The dyeing processes form an integral part of textile industry which uses 1.3 million tons of dyes and dye precursors.7 About 10,000 different synthetic dyes are used in textile dyeing industries with over 7×105 tons of these dyes produced annually worldwide.8,9 Nearly 10-15% of these dyes does not bind to fabric and is released into the environment during the dyeing process.10-12 Since these dyes are harmful their discharge poses serious environmental problems.13 The important classes of synthetic dyes include anthraquinone, triaryl methane and azo dyes. Azo dyes are the largest group of organic synthetic dyes extensively used in paper making, cosmetic and textile industries.14,15 The major group of dyes mostly used in industry are azo dyes.16-18 These are characterized by the presence of one or more azo groups (-N=N-). As per estimates nearly 2,000 different azo dyes are used to dye various materials such as textiles, leather, plastics, cosmetics and food. A number of physicochemical methods are available for the remediation of dye wastewaters such as adsorption, chemical oxidation, precipitation, coagulation, filtration, electrolysis and photodegradation. The major disadvantages of these techniques are high cost, low efficiency, limited versatility, interference by other wastewater constituents and secondary waste generation.19,20 Remediation using biological methods is generally considered environmentally friendly as it leads to complete mineralization of organic pollutants at low cost.21 Use of microorganisms offers considerable advantages in being relatively inexpensive, low running cost and the end products of mineralization are not toxic.22 Bacterial decolorization of dyes has gained importance in recent times and is found to be normally faster compared to fungal decolorization.7,23-25 The current study aims to evaluate the ability of the strain Bacillus mojavensis ABR in decolorizing and degrading the azo dye Direct Orange 26 (DO-26). The strain ABR was assessed for its potential to decolorize DO-26 at different pH, temperature, salinity and increasing dye concentration under static conditions. DO-26 degraded products were analysed by High Pressure Liquid Chromatography (HPLC) and Liquid Chromatography Mass Spectrometer (LC-MS). The phytotoxicity study on the seeds of Vigna radiata, Sorghum bicolor and Vigna mungo revealed the less toxic nature of the metabolites.
MATERIALS AND METHODS
Chemicals
The Azo dye Direct Orange 26 (DO-26) with maximum absorption at 493 nm (λmax) was obtained from a local textile mill in Tamil Nadu, India in pure form. HPLC- grade methanol was purchased from Sigma. Dye stock solution was prepared by dissolving 1000 mg L-1 of DO-26 in double distilled water and purified by membrane filtration (0.22 µm). The structure of the dye is shown in Fig. 1.
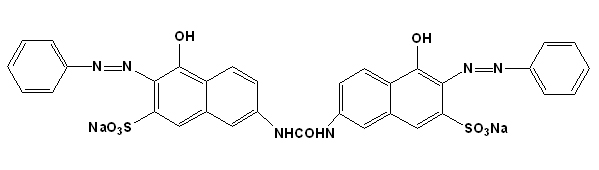
Figure 1. Chemical structure of the Azo dye Direct Orange 26.
Analytical Methods
UV-Visible spectrophotometer (Hitachi U-2800) was used to analyse decolorization. Biodegradation was assayed by HPLC (Waters, Model No.501) and HPLC-MS (Thermo Finnigan Surveyor-Thermo LCQ Deca XP MAX).
Isolation, Screening and Identification of Dye decolorizing Bacteria from Textile Effluent
The collected textile effluent from Coimbatore was serially diluted from 10-1 to 10-7and plated on Luria Bertani (LB) Agar plates and were incubated at 37°C for 24h. Distinct colonies were separated and repeatedly streaked on LB Agar plates to obtain pure colonies. Initial screening for DO-26 decolorization was carried out in 50 ml of decolorizing media containing gl-1 Peptone, 10, Meat Extract, 10, NaCl, 5 under static conditions with 5 mg L-1 of DO-26. 5 mg L-1 of DO-26 was added to the overnight grown isolates and incubated under static conditions to check for decolorization. The isolate which exhibited highest potential in decolorizing DO-26 was chosen and identified by molecular method using 16S rRNA gene sequencing.26 The obtained gene sequence was evaluated at NCBI server (http://www.ncbi.nlm.nih.gov) using BLAST (blastn) tool and the homology was analysed using CLUSTAL W.27 The sequences were aligned by the neighbour-joining method using Kimura-2-parameter distances and phylogenetic tree was constructed in MEGA version 5 package.28 The nucleotide sequence data obtained was deposited in the GenBank under accession number HF568798.
Dye decolorization study
A loopfull of the potential isolate (Bacillus mojavensis ABR) was inoculated in LB broth and precultured with shaking at 130 rpm for overnight.29 7% (v/v) of the pregrown inoculum of the potential isolate was used to inoculate 100 ml of decolorizing medium in 250 ml Erlenmeyer flasks containing 100 mg l-1 of Direct Orange 26 dye. The process of decolorization was carried out by incubating the set up at pH 7 and 37°C under static conditions. At regular intervals (0hr, 2hr, 4h and 6hr) an aliquot (3ml) of sample was withdrawn and centrifuged at 8,000 × g for 15 min. The supernatant was separated and assayed for decolorization by measuring the absorbance at 493 nm (λmax). Control (uninoculated medium) was always included. The percentage of reduction in dye concentration was analysed by the formula: (%) = Ci-Cr/Ci×100%, where Ci and Cr were initial and residual dye concentrations, respectively. All experiments were carried out in triplicate.
Effect of external factors on DO-26 decolorization
The effect of external factors on DO-26 decolorization by B. mojavensis ABR was assayed. The effect of various pH (6, 7, 8 and 9), temperatures (32, 37, 40 and 45°C) and salinity levels (2, 5, 7 and 10% NaCl) was analysed in decolorizing medium in 250 ml Erlenmeyer flasks containing 100 mg l-1 of Direct Orange 26 dye under static conditions.
Degradation Analysis by HPLC and LCMS
The analysis of degraded metabolites of Direct Orange 26 (DO-26) by B. mojavensis ABR was carried out by High Pressure Liquid Chromatography (HPLC) and Liquid Chromatography Mass Spectrometer (LC-MS). The culture broth (100 ml) was withdrawn at initial and final hours (before and after decolorization), centrifuged at 10, 000g for 15 min and metabolites were extracted from the supernatant using equal volumes of ethyl acetate. The extracts were dried and evaporated to dryness in rotary evaporator. Degraded metabolites were analysed by HPLC on reverse phase C18 column (symmetry, 4.6×150 mm) equipped with dual λ UV-Vis detector. The collected samples at different time (0h and 6h) were dried and dissolved in 2 mL HPLC grade methanol. The mobile phase used was methanol/water (50:50, v/v) with a flow rate of 0.8 mL min-1 for 10 min and UV detector set 493 nm. To analyze the dye degradation, the HPLC-MS (Thermo Finnigan Surveyor - Thermo LCQ Deca XP MAX) with Xcalibur software was used. 10 µL of the prepared solutions containing degraded dye moieties were injected into the LC/MS and the mass spectra of the samples were then determined. The composition of the mobile phase was a mixed solution of water and methanol (50:50) and its flow rate was 0.2 mL/min for 60 min run time. The stationary phase was the C18 reverse phase column (BDS HYPERSIL C18, 4.6 × 250 mm, 5.0 µm). The mass spectra were obtained using Electro Sprat Ionization (ESI) under the flow of Helium gas at 1mL/min approximately and the fragment voltage was 16 (V).
Phytotoxicity Study
The seeds of V. radiata, S. bicolor and V. mungo were used for phytotoxicity study against 100 ppm concentration of DO-26 dye and its degraded metabolites. The seeds of respective plants were grown with fixed volume of azo dye (DO-26), degraded metabolites and distilled water as control. The difference in root in shoot lengths of these seeds was measured after a period of seven days.
Statistical Analysis
All the experiments were carried out in triplicate and the data was analysed using ANOVA with Tukey-Kramer multiple comparison test. The results were expressed as the mean ± SD (standard deviation).
Figure 2. Bacillus mojavensis strain ABR pure culture on Luria Bertani Agar.
RESULTS AND DISCUSSION
Isolation, Screening and Identification of DO-26 decolorizing bacteria from Textile Effluent
Out of 15 bacterial isolates from the textile effluent sample from Coimbatore, the potential bacterial strain Bacillus mojavensis ABR (Fig. 2) was chosen based on its ability to decolorize Direct Orange 26 (DO-26) in decolorizing medium. 100 mg L-1 of DO-26 azo dye was decolorized by strain ABR with 95% rate at 37 °C, pH 7 and in the presence of 7% NaCl under static conditions in 6 h. Based on 16S rRNA sequence comparison the active isolate was identified as Bacillus mojavensis strain ABR (1341 bp). The homology study indicated that the strain ABR had 98% nucleotide identity with Bacillus mojavensis strain LZ013 (JQ073762). The relationship of strain ABR to other Bacillus species is shown in a phylogenetic tree depicted in Fig. 3.
Decolorization of DO-26 by Bacillus mojavensis strain ABR
The decolorization of DO 26 azo dye by Bacillus mojavensis was shown in Fig. 4.
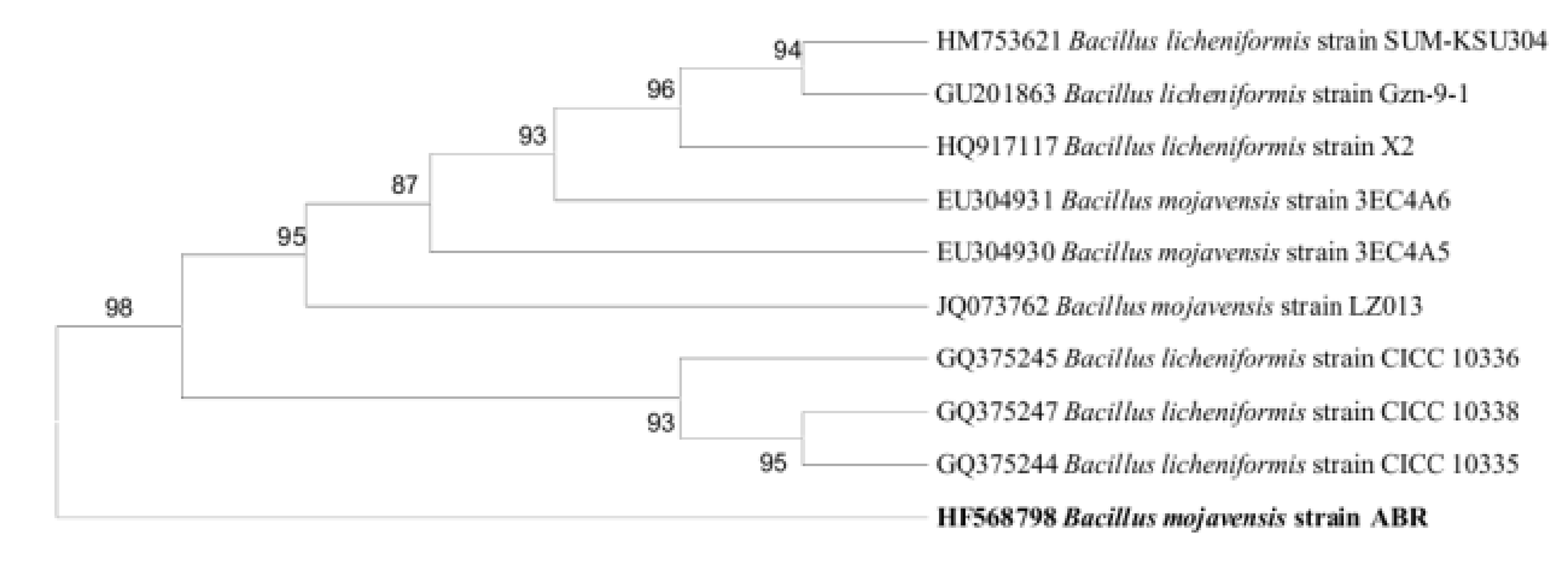
Figure 3. Phylogenetic tree of Bacillus mojavensis strain ABR.

Figure 4. Erlenmeyer flasks showing decolorization of DO-26 by Bacillus mojavensis strain ABR a.) Control b.) DO-26 decolorized ( 6 hrs) c.) DO-26 (0 hrs).
The result of azo dye (DO-26) decolorization by Bacillus mojavensis strain ABR by UV-Vis spectrophotometer is depicted in Fig. 5. The initial dye peak at 493 nm was found to decrease significantly with decolorization at the end of 6 h. Bacillus mojavensis strain ABR decolorized DO-26 at a concentration of 100 mg L-1 by 95% efficiency within a time span of 6 h at 37°C and pH 7 under static conditions.
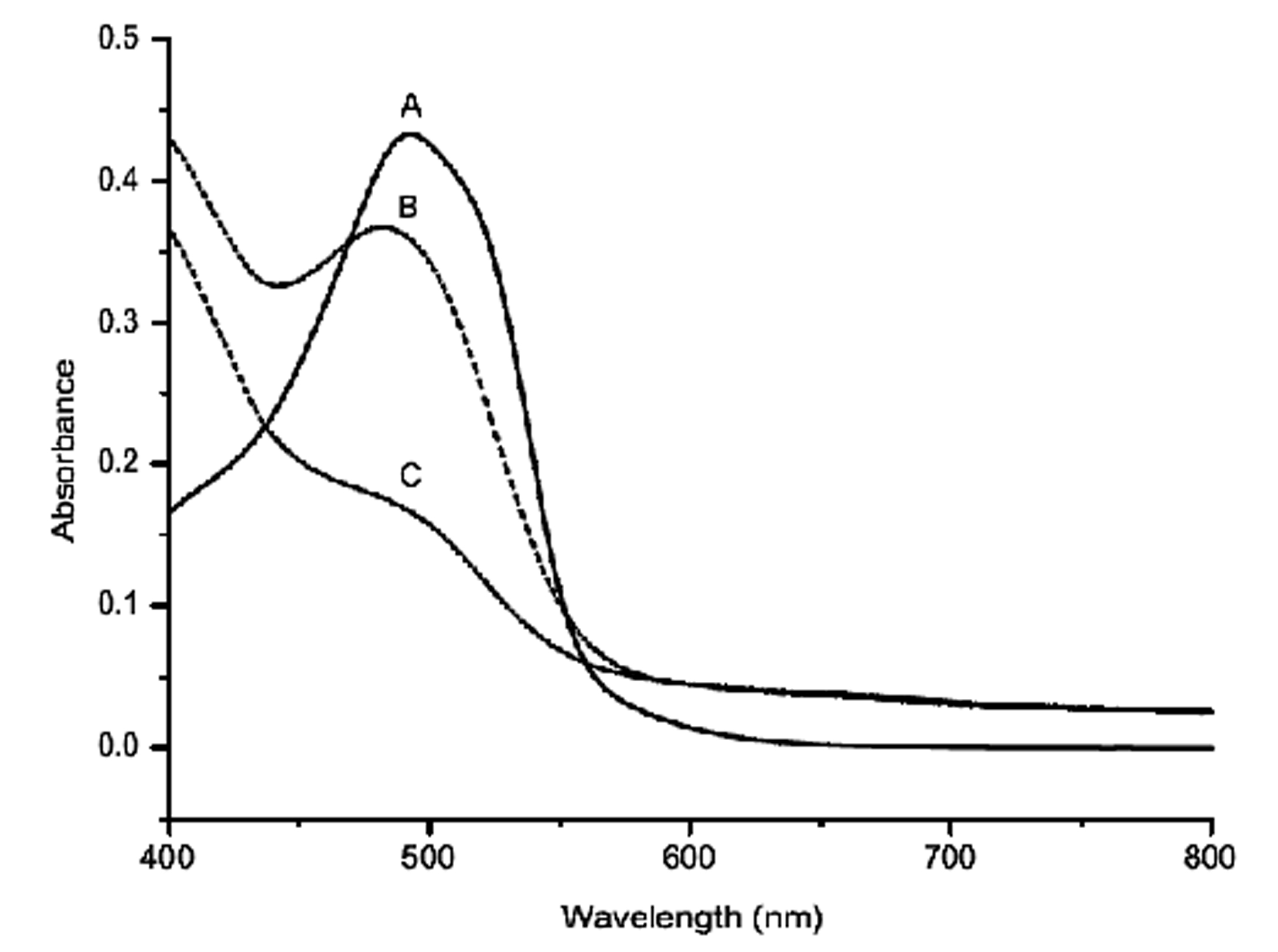
Figure 5. UV- Scan of supernatant extracted at different time intervals during DO-26 decolorization. Samples withdrawn at 0hr, 4hr and 6hr (A: 0hr spectra; B: 4h spectra; C: 6hr spectra).
Effect of external parameters on DO-26 decolorization
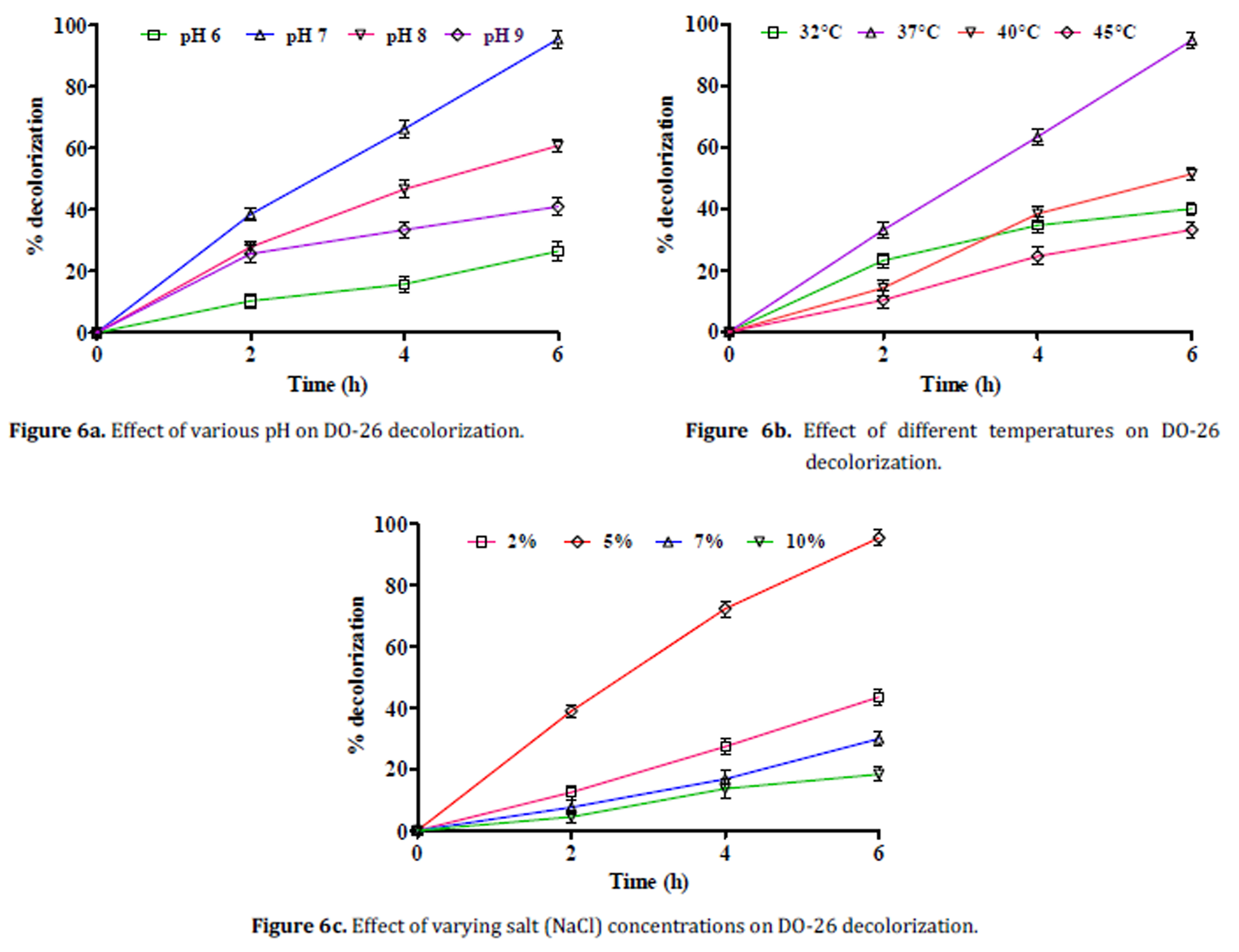
Environmental factors are known to play a key role in the process of decolorization of dyes by microorganisms.30 Bacillus mojavensis ABR decolorized DO-26 dye with 95% efficiency at pH 7 under static conditions (Fig. 6a). Decolorization rates of about 26, 60 and 41% were noticed at pH 6, 8 and 9 respectively. The effect of temperature on DO-26 decolorization by ABR is represented in Fig. 6b. The optimal temperature for DO-26 decolorization was 37°C. Increase in temperature resulted in gradual decrease in decolorization rate exhibiting 51% and 33% at 40°C and 45°C. Decolorization rate was found to be 40% at 32°C. Similar results were observed with respect to temperature in Bacillus cohnii MTCC 3616 and Deinococcus radiodurans R1.31,32 Decreased rate of decolorization at high temperatures could be due to deactivation of enzymes.33,34 DO-26 decolorization in the presence of varying salt concentrations is depicted in the Fig. 6c. The rate of DO-26 decolorization increased with NaCl concentration. Optimal decolorization (95%) of DO-26 by B. mojavensis ABR was observed at 5% NaCl concentration (Fig. 4c). At 2% and 7% NaCl concentrations the rates of decolorization was 43% and 30% respectively. Lowest rate of decolorization of about 18% was noticed at higher (10%) NaCl concentration. The reason for decreased decolorization rate at higher salt concentrations is due to stress as bacterial cells cannot tolerate high salt concentrations.35 Similar inhibition activity at high salt levels was observed in Bacillus sp. YZU1 decolorizing Reactive Black 536 and in Bacillus cohnii MTCC 3616.31
DO-26 biodegradation analysis by HPLC and LCMS
The result of HPLC analysis of azo dye Direct Orange 26 (DO-26) is depicted in Fig. 7a. The dye (DO-26) eluted as a single peak at a retention time of 2.98 min. The single peak of the dye represents its purity.
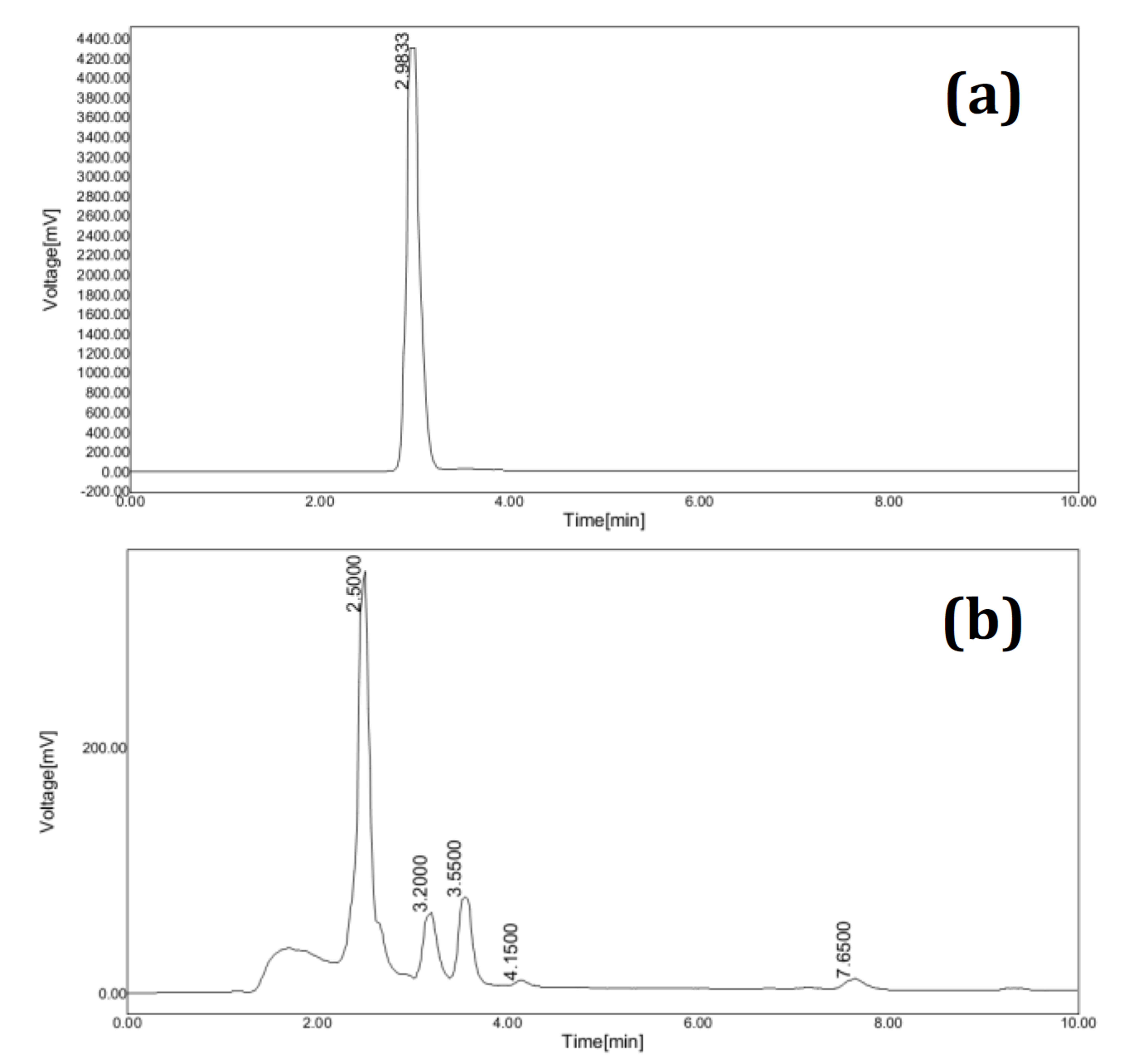
Figure 7. (a) HPLC chromatogram of Direct Orange 26 extracted at 0hr. (b) HPLC elution profile of degraded metabolites extracted at 6hr.
The HPLC chromatogram after azo dye degradation showed a major peak at 2.50 min and minor multiple peaks were observed at 3.20, 3.55, 4.15 and 7.65 min respectively. HPLC chromatogram of biodegraded sample is shown in Fig. 7b. Biodegradation of DO-26 by B. mojavensis strain ABR is indicated by the absence of dye peak and the emergence of multiple peaks at different retention times.
Table 1. Phytotoxicity comparison of DO-26 and its extracted metabolites.
|
Parameters |
Vigna mungo |
Sorghum bicolor |
Vigna radiata |
|
||||||||||||||
|
Water |
DO-26 (100 ppm) |
Extracted metabolites (100 ppm) |
Water |
DO-26 (100 ppm) |
Extracted metabolites (100 ppm) |
Water |
DO-26 (100 ppm) |
Extracted metabolites (100 ppm) |
|
|||||||||
|
Germination (%) |
100 |
30 |
100 |
100 |
20 |
100 |
100 |
20 |
100 |
|||||||||
|
Shoot (cm) |
9.02±0.13 |
2.13±0.2** |
7.14±0.47* |
8.66±0.69 |
2.14±0.14** |
6.34±0.69* |
10.23±0.56 |
1.89±0.61** |
7.36±0.34 |
|||||||||
|
Root (cm) |
3.04±0.15 |
00.35±0.13** |
2.04±0.20 |
3.46±0.11 |
0.62±0.12** |
2.50±0.19 |
4.12±0.19 |
0.88±0.13** |
2.40±0.34* |
|||||||||
Values are mean of three experiments ± SEM, significantly different from the control (seeds germinated in water) at *P< 0.05, **P< 0.01, by one-way analysis of variance (ANOVA) with Tukey–Kramer comparison test.
The purpose of LC-MS analysis is to identify the intermediates produced from the degradation process. The degradation of DO-26 dye is clearly confirmed by comparing the LC-MS chromatograms of DO-26 dye and its degraded sample which are represented in Fig. S1 and Fig. S2 (see supporting information). LC-MS analysis of the dye and their degraded product confirmed degradation of Direct Orange 26 by Bacillus mojavensis ABR. The retention times of all degradation products are in the range of 8.61 – 13.65 which are different from the retention time of control dye Direct Orange 26 at 6.71. These data clearly show the biodegradation of Direct Orange 26 azo dye by Bacillus mojavensis strain ABR.
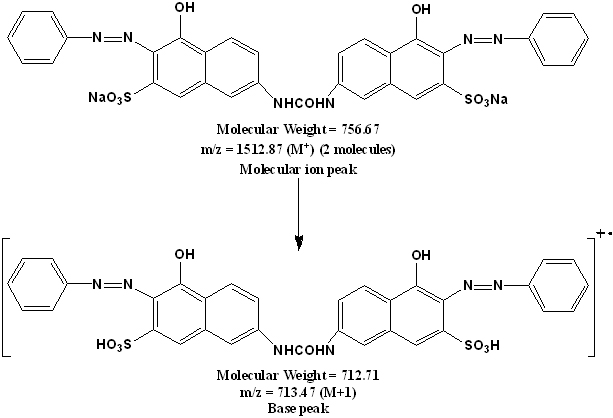
Figure 8. Proposed mass fragmentation pattern of Direct Orange 26 dye.
The chemical structure of the Direct Orange 26 dye (Fig. 1) is confirmed through LC-MS spectral analysis. Direct Orange 26 dye showed a single peak in the LC-MS chromatogram at a retention time of 6.71 min. The molecular ion peak appeared at m/z 1512.87 (for two molecules) and the base appeared peak at m/z 713.47 in the LC-MS spectrum which confirms the chemical structure of the Direct Orange 26 dye (Fig. 8).
The peak present in the LC-MS chromatogram of the DO-26 dye was absent in the chromatogram of degraded products and the retention time of degraded products are in the range of 8.61-13.65 minutes which are entirely different from the retention time of DO-26 dye which is at 6.71 min. This significant change in the retention times indicates that the azo dye (DO-26) has been degraded to low molecular fragments by the strain ABR. Furthermore, the high intense peaks in the LC-MS spectra of Direct Orange 26 dye degraded product with molecular mass of 453, 471, 227 and 279 shows a prominent deviation when compared with the high intense molecular masses (713 and 735) in the LC-MS spectrum of DO-26 dye. The difference in the retention time of peaks of Direct Orange 26 dye and its degraded product in the LC-MS chromatographs and the fragmented peaks in the LC-MS spectra confirms the degradation of Direct Orange 26 dye by Bacillus mojavensis strain ABR.
Phytotoxicity Study
The untreated dyeing effluents have direct and serious effects on the environment as well as soil fertility. The seeds of V. mungo, S. bicolor and V. radiate were tested against the azo dye Direct Orange 26 and its degraded metabolites at a concentration of 100 ppm. DO-26 treated seeds exhibited low germination percentage in V. mungo (30%), S. bicolor (20%) and V. radiata (20%). 100% germination rate was recorded in all three seeds when treated with 100 ppm concentration of degraded metabolites of DO-26 and distilled water. The azo dye DO-26 was found to significantly affect the root and shoot length of the seeds compared to the metabolites after decolorization and degradation (Table 1). The phytotoxicity study clearly indicates the potential of Bacillus mojavensis strain ABR in degrading and detoxifying the azo dye Direct Orange 26 efficiently.
Conclusions
The present study determines the potential of Bacillus mojavensis strain ABR isolated from textile effluent to decolorize and degrade the azo dye Direct Orange 26 (DO-26). The acclimatized strain decolorized 100 mg L-1 DO-26 with 95% efficiency at pH 7, 37°C and in the presence of 5% NaCl within a short time span of 6 h. The strain ABR was able to degrade the dye DO-26 as detected by HPLC and LCMS by the formation of metabolites. Bacillus mojavensis ABR led to the successful detoxification of DO-26 being proved by the phytotoxicity test. Hence, Bacillus mojavensis strain ABR could be practically employed in decolorizing and degrading several of the complex Azo dyes and to effectively treat dyeing effluents.
Acknowledgements
The authors acknowledges the management of the VIT University, Vellore for providing necessary facilities. The authors would also like to acknowledge the assistance of Dr. V. S. V. Satyanarayana, IIT Mandi.
Notes and References
* Corresponding Author Details: A. S. Arun Prasad, Division of Environmental Biotechnology, School of Biosciences and Technology, VIT University, Vellore, India.
E-mail id.: This email address is being protected from spambots. You need JavaScript enabled to view it.
† Supporting Information (SI) available: [LC-MS Spectra].
1. I. O. Asia, N. A. Oladoja and E. E. Bamuza-pemu, African Journal of Biotechnology, 2006, 5, 1678.
2. G. Tchobanoglous and F. L. Burton, Wastewater Engineering: Treatment, Disposal and Reuse/Metcalf and Eddy, (Tata McGraw-Hill Publishing Company Limited: New Delhi, India), 1995.
3. B. S. Andre dos, J. C. Francisco and B. L. Jules van, Bioresource Technol., 2007, 98, 2369.
4. A. M. Talarposhti, T. Donnelly and G. K. Anderson, Water Res., 2001, 35, 425.
5. A. B. Dos Santos, I. A. E. Bisschops and F. J. Cervantes, Closing process water cycles and product recovery in textile industry: perspective for biological treatment. F. J. Cervantes, A. C. Van Haandel and S. G. Pavlostathis (ed.) (Advanced Biological Treatment Processes for Industrial Wastewaters, Vol. 1. International Water Association: London), 2006, p. 298.
6. Environmental Protection Agency. Profile of the Textile Industry (Washington: EPA), 1997.
7. J. S. Chang, C. Chou and S. Y. Chen, Water Sci. Technol., 2001, 43, 261.
8. U. Meyer, Biodegradation of synthetic organic colorants. Microbial degradation of xenobiotic and recalcitrant compounds. T. Leisinger, A. M. Cook, R. Hunter, J. Nuesch, (ed.) (FEMS symposium 12, Academic Press: London), 1981, p. 371.
9. H. Zollinger, Colour Chemistry-Synthesis, Properties of Organic Dyes and Pigments, (VCH Publishers: New York, USA), 1987, p. 92.
10. J. T. Spadary, L. Isebelle and V. Renganathan, Indian J. Biotech., 1994, 1, 393.
11. F. He, W. Hu and Y. Li, Chemosphere, 2004, 57, 293.
12. M. S. Reish, Chem. Eng. News Jan., 1996, 15, 10.
13. P. Verma and D. Madamwar, World J. Microbiol. Biotech., 2003, 19, 615.
14. I. M. Banat, P. Nigam, D. Singh and R. Marchant, Bioresource Technol., 1996, 58, 217S.
15. T. Robinson, G. McMullan, R. Marchant and P. Nigam, Bioresource Technol., 2000, 77, 247.
16. H. Chen, R. F. Wang and C. E. Cerniglia, Protein Exp. Purif., 2004, 34, 302.
17. K. Kumar, S. S. Devi, K. Krishnamurthi, S. Gampawar, N. Mishra, G. H. Pandya and T. Chakrabarti, Bioresource Technol., 2006, 97, 407.
18. J. P. Jadhav, G. K. Parshetti, S. D. Kalme and S. P. Govindwar, Chemosphere, 2007, 68, 394.
19. F. P. Van der Zee and S. Villaverde, Water Res., 2005, 39, 1425.
20. P. Kaushik and A. Malik, Environ. Int., 2009, 35, 127.
21. A. Pandey, P. Singh and L. Iyengar, Int. Biodeter. Biodegrad., 2007, 59, 73.
22. E. Forgacs, T. Cserhati and G. Oros, Environ. Int., 2004, 30, 953.
23. M. F. Coughlin, B. K. Kinkle and P. L. Bishop, Chemosphere, 2002, 46, 11.
24. G. Buitron, M. Quezada and G. Moreno, Biores.Technol., 2004, 92, 143.
25. K. Sarayu and S. Sandhya, Appl. Biochem. Biotechnol., 2010, 160, 1241.
26. S. F. Attschul, T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller and D. J. Lipman, Nucleic Acids Res., 1997, 25, 3389.
27. J. D. Thompson, T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, Nucleic Acids Res., 1997, 24, 4876.
28. K. Tamura, D. Peterson, N. Peterson, G. Stecher, M. Nei and S. Kumar, Mol. Biol. Evol., 2011, May 4, 1.
29. J. Sambrook, E. F. Fritsch and T. Maniatis, Molecular cloning: a laboratory manual (2nd ed.) (Cold Spring Harbor Laboratory, Cold Spring Harbor: NY), 1999.
30. C. I. Pearcea, J. R. Lloydb and J. T. Guthriea, Dyes Pigments, 2003, 58, 179.
31. A. S. Arun Prasad and K. V. Bhaskara Rao, Appl. Microbiol. Biotechnol., 2013, 97, 7469.
32. G.Y. Lv, J. H. Cheng, X. Y. Chen, Z. F. Zhang and L. F. Fan, Bioresour. Technol., 2013, 144, 275.
33. D. Cetin and G. Donmez, Enzyme Microb. Tech., 2006, 38, 926.
34. T. Panswad and W. Luangdilok, Water Res., 2000, 34, 4177.
35. A. Zilly, C. M. J. Da Silva, A. Bracht, C. G. Marques de Souza, A. E. Carvajal, E. A. Koehnlein and R. M. Peralta, Int. Biodeter. Biodegrad., 2011, 65, 340.
36. Z. W. Wang, J. S. Liang and Y. Liang, Int. Biodeter. Biodegrad., 2013, 76, 41.
